Expand Condon
Because the the using of D-Alawe need to find a way that can synthesize protein with D-Ala.We decide to introduce D-Ala into the bacteria translation system.
In the translation process,a kind of amino acid is caught by a specific aminoacyl-tRNA synthetase (aaRS).The aaRS recognise the corresponding tRNA and connect the tRNA and amino acid together.The amino acid is brought to the activated set in ribosome with the identification of codon on mRNA and anti-codon on tRNA.There is 20 similar paths in cells ,which have nothing to do with each other.[1]

We need to generate a orthogonal path to introduce D-Ala, and All we need is a unused codon ,a corresponding tRNA ,and an aaRS that can recognise D-Ala and the new tRNA.[1]
It is relatively easy to get a codon and tRNA.We will use the amber asthe codon of the D-Ala,which is hardly used in E.coli. We choose a tRNAAlafrom Aquifex aeolicus to make sure it won't disturb the other path in E. coli ,and change its anti-codon to correspond amber.

The most difficult part is to designa D-AlaRS.We use the AlaRS also from Aquifex aeolicus to mutate.Ala and Gly are similar with D-Ala, so we will analysis the intereation of AlaRS with Ala and Gly[2].
The electropositive amino group is stabilized by interaction with the invariant motif 3 Asp-217 side chain carboxyl.
One of the carboxylate oxygen atoms of the amino acid substrate makes a salt bridge (water-mediated in the case of Gly) with the side chain of invariant Arg-69 of motif 2.
A water-mediated hydrogen bond between the α-amino group of bound amino acids and the NH of Trp-161.
A hydrogen bond donated by the δ-amine of Asn-194 to the other carboxylate oxygen atom of the bound amino acid.
AInvariant Met-92 (from motif 2), highly conserved Val-196 (fromstrand 10) and Gly-219 (from motif 3) line the pocket from the bottom, top, and side, respectively, and make van der Waals contacts with the methyl group of Ala (Fig. 2B Top). Gly fits in this pocket and sits deeper in the pocket to make a contact with Met-92 by means of its C atom.
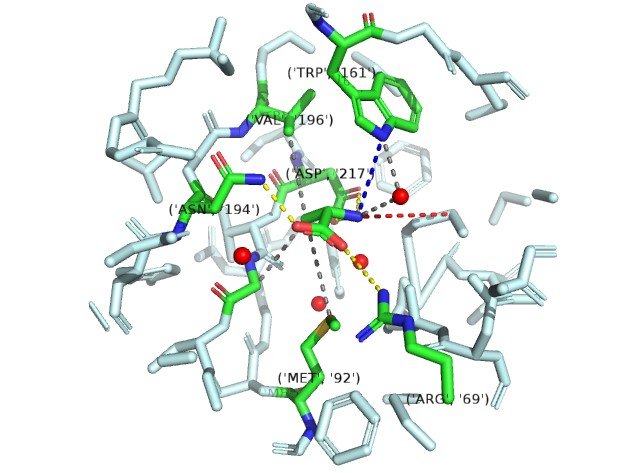
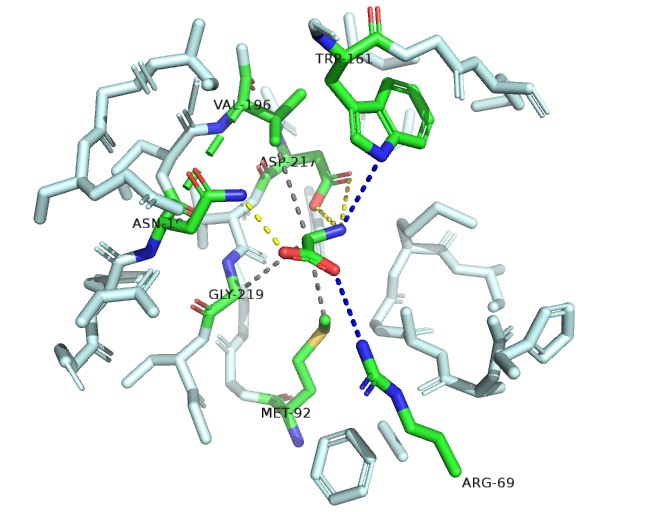
After analysising those intereation and selecting the residue by the distance between residue and binding point ,We decided to have a mutation: Asp217->Glu Trp161->Tyr 缺模拟数据

[1] Nediljko Budisa Prolegomena to Future Experimental Efforts on Genetic Code Engineering by Expanding Its Amino Acid Repertoire Angew. Chem. Int. Ed. 2004, 43, 6426 – 6463
[2] Manal A. Swairjo and Paul R. Schimmel Breaking sieve for steric exclusion of a noncognate amino acid from active site of a tRNA synthetase PNAS January 25, 2005 vol. 102

